 Evolution & Behaviour
Evolution & Behaviour
When it comes to the giant bacterium Achromatium, everything is everywhere
Unlike typical bacteria, the giant Achromatium contains hundreds of unidentical chromosomes. Achromatium is present globally in different environments. This typically leads to speciation. Nevertheless, Achromatium shows minimal environment-specific phylogenetic differentiation. Instead, it harbors a globally identical functional inventory for which uses genes it needs for the specific habitat.
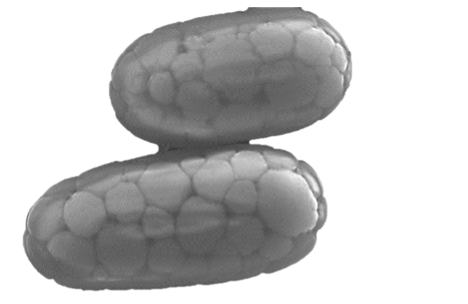
Bacteria of the genus Achromatium are known since the 1890s. They occupy the uppermost layer of aquatic sediments at the boundary between the oxic water and the anoxic benthos where Achromatium uses sulfide for energy. Achromatium is recognizable by two distinctive features: (1) it is a giant bacterium with a volume 30,000 times larger than any typical bacteria, (2) it harbors “stones” (made of calcium carbonate) that fill most of its cell body.
Like other giant bacteria, Achromatium contains many chromosomes on which its genetic material is encoded, typically around 300 chromosomes. However, unlike anything seen so far among bacteria, these chromosomes are not identical, making Achromatium the only known bacterium with multiple different chromosomes having numerous different versions of all functional genes including those known to occur only once in a bacterial genome.
We believe that this unprecedented genomic diversity is tightly linked to the stones in the cell. The cytoplasm is “squeezed” in between stones forming a thin network with occasional larger compartments. These compartments are likely isolated one from another and so are the clusters of chromosomes trapped in them. Though these chromosome clusters had a common ancestor, they evolve independently from one another, giving rise to multiple alleles of the same gene in one single cell. When cells divide, chromosome clusters close to the division plane may mix, resulting in new genomic combinations. If the latter proves right, it is a single-cell bacterial representation of sexual-like reproduction, known thus far solely from Achromatium.
For more than a century, Achromatium was found exclusively in freshwater, known as the largest freshwater bacterium. Recently, however, it was also described from several saline environments. Therefore, we set to investigate the global distribution and diversity of the genus Achromatium.
To do so, we screened all publicly available DNA data obtained from sediments, for genes belonging to Achromatium. Achromatium was found to be universal, inhabiting a broad range of environments from the Arctic to the Antarctic. It occurs in shallow waters as well as in the ocean at depths of 4000 meters. It was found in hot springs but also in ice-cold water, in acidic and alkaline sediments, and also in hypersaline waters.
Typically, such a wide range of environmental conditions results in the establishment of new species, well-adapted to their specific environment. However, Achromatium defies this expectation. When comparing the sequences of phylogenetic markers or functional genes, no habitat-specific separation was observed. This is in contrast for example to the Pelagibacter clade, the most abundant group of aquatic bacteria, in which freshwater and marine species form separate phylogenetic groups.
Even more surprising was the lack of any functional difference between Achromatium-like sequences recovered from different environments. In a process called genome streamlining, bacteria typically adapt to their habitat, losing unnecessary genes. This seems not to be the case for Achromatium and two results point to the possibility of an evolutionary strategy previously unknown. First, though the overall functional inventory was identical across environments, our data point to a difference in the copy number of genes among cells from different habitats. Second, when we investigated transcriptomic data, showing which genes of the total inventory are used, clear patterns were observed separating different habitats.
Thus, we propose that Achromatium accumulates functions with time. It adapts to different habitats not by losing unnecessary genes but rather by changing their copy numbers, enriching those it needs and “archiving” those it does not. Thereafter, Achromatium appears to regulate which genes it uses based on what it needs to survive in its current habitat. This previously undescribed strategy explains the ability of Achromatium to exist in environments with drastically different characteristics and suggests that it possesses the ability to overcome dispersal limitation by adapting each time to any environment it encounters.
Our understanding of Achromatium so far, consisting of its, likely, compartmentalized internal structure, and continuous shuffling of chromosomes, similar to sexual reproduction for the generation of genomic diversity and its evolutionary strategy of function accumulation and archiving, points to an organism with bacterial metabolism featuring many characteristics of advanced unicellular and multicellular organisms. However, whether Achromatium served a role in the evolution of sexual reproduction and the development of multicellular organisms or whether it is a unique bacterium with a special capability for adaptation remains to be further determined.
The authors would like to thank the MIBI group at IGB for fruitful discussions and Artur Zaduryan, Mina Bizic, Solvig Pinnow, Hans-Peter Grossart and Heribert Cypionka for their valuable contribution to the original publication.
Original Article:
Ionescu, D. et al. Heterozygous, Polyploid, Giant Bacterium, Achromatium, Possesses an Identical Functional Inventory Worldwide across Drastically Different Ecosystems. Molecular Biology and Evolution 38, 1040-1059 (2020).Edited by:
Isa Ozdemir , Senior Scientific Editor
We thought you might like
Natural products might just be our best weapon against antibiotic resistance
Apr 3, 2024 in Maths, Physics & Chemistry | 3.5 min read by Olivier Kirchhoffer , Jahn Nitschke , Jean-Luc WolfenderResponding to sea-level rise: the importance of culture
Jun 10, 2021 in Earth & Space | 4 min read by Robert L. Barnett , Sophie L. WardWarm waters hide in the unlikeliest of places – under the Arctic sea ice
Jul 31, 2019 in Earth & Space | 3.5 min read by Mary-Louise TimmermansFancy footwork: Darwin’s pigeons and the evolution of foot feathers
Jan 20, 2017 in Evolution & Behaviour | 3.5 min read by Eric DomyanMore from Evolution & Behaviour
Cicada emergence alters forest food webs
Jan 31, 2025 in Evolution & Behaviour | 3.5 min read by Martha Weiss , John LillSize does not matter: direct estimations of mutation rates in baleen whales
Jan 29, 2025 in Evolution & Behaviour | 4 min read by Marcos Suárez-MenéndezThe Claws and the Spear: New Evidence of Neanderthal-Cave Lion Interactions
Jan 22, 2025 in Evolution & Behaviour | 3.5 min read by Gabriele RussoA deep-sea spa: the key to the pearl octopus’ success
Jan 20, 2025 in Evolution & Behaviour | 3.5 min read by Jim BarryFeisty fish and birds with attitude: Why does evolution not lead to identical individuals?
Aug 31, 2024 in Evolution & Behaviour | 3 min read by Lukas Eigentler , Klaus Reinhold , David KikuchiEditor's picks
Trending now
Popular topics


