 Evolution & Behaviour
Evolution & Behaviour
Coronaviruses: Contagious Beasts and Where to Find Them
Analysis of the CoV-19 genome reveals two crucial mutations responsible for its efficient capacity to infect humans. There are two possible scenarios of how these mutations could have emerged. Both of them dismiss the possibility that the CoV-19 is a human-made virus that escaped from some laboratory.
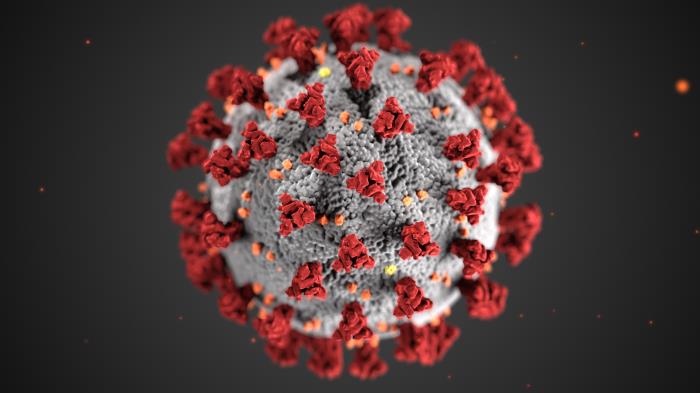
First and foremost: viruses do not appear out of nowhere. They exist in nature moving from one hosting animal to another. Still, we don't realize their presence until they cross our way. For example, the first time a coronavirus was discovered was back in the 1930s in domestic chickens. That coronavirus is a distant relative of the CoV-19, but they are not the same.
Viruses are bizarre things. They cannot be classified as alive since they cannot live and reproduce autonomously. Instead, these little clumps of genetic code (DNA or RNA strands), encapsulated into a special coating, break into cells of others and use them to procreate. Each virus has its particular specification: it can infect only certain types of tissues in certain animal species. For example, there is a receptor protein called ACE2 that sits on the surface of our lung cells and regulates the blood pressure. This unfortunate protein turned out to be a target for the CoV-19, but also for equally notorious SARS-related coronavirus (SARS-CoV) that caused an epidemic in 2002-2003. The lungs are the primary victims of these infections because ACE2 is their entrance door.
Viruses tend to be "faithful" to their host-species, but sometimes a so-called spillover happens. That is when a virus becomes able to infect new species. CoV-19 is already the seventh coronavirus known to infect humans, but only three of these seven, including CoV-19, can cause severe symptoms.
To understand how the virus "learns", we need to understand what it is. Structurally speaking, the virus is literally a thing in itself. Within its core, there is a molecule of nucleic acid (RNA in coronaviruses) that bears a genetic code for a few viral proteins. The information embedded in the genetic code leads to the assembly of spike proteins that sit on the surface of the virus' body. These spike proteins are the key for the virus's infection, as they can bind receptors (the lock) on the host cells (like mentioned ACE2), pretending that they are harmless friends. Yet, this trick-or-treating game turns into a disaster when the virus opens the door and injects its genome into the target cell. Once inside, the nucleic acid hijacks the reproducing machinery and it creates a lot of copies of viral genome to produce many more viral proteins. With this, new copies of viruses assemble and literally break out of the cell, ready to infect new cells.
The key to the success of the virus is the affinity between its spike proteins and their targets located on the cells that will become infected.
Like everything in nature, this affinity is a subject of evolution, which as ever happens through random tinkering of the viral genome. Substitutions, deletions or insertions of its main components, the nucleotides, happen here just like in any other genome. And some of these changes lead to improved or entirely new affinities. That is how influenza "strikes back" every winter season. That is also how some viruses jump from one species to another.
The difference between the coronavirus strains recently isolated from bats, pangolins, and human CoV-19 is minimal but essential. It seems to boil down to two mutations within the genetic code of the spike proteins: a few substitutions in the binding domain and a short insertion in another, cleavage, site of the protein. These types of changes are not unique and were also found in other viruses from the same family. That is strong evidence that these changes are evolutionary natural to occur. The CoV-19 substitutions account for its affinity to the ACE2 receptor and the insertions drastically improve its capacity to get into a cell.
According to this recent article published by Andersen and colleagues, the two-fold difference of CoV-19 from its closest known relatives suggests two possibilities of its emergence. Either infective insertion occurred before the virus broke the barrier between original hosts and humans; or, first CoV-19 learned to inefficiently infect humans and then acquired its highly efficient infective genomic insertion through an unregistered human-to-human exchange. The fact that the coronavirus that infects pangolin has a binding domain almost identical to the CoV-19 makes it probable that the virus first settled in humans and then learned how to infect efficiently. In this case, future retrospective analysis of human serum samples might discover the previous, inefficient, version of CoV-19.
In any case, the third scenario, where CoV-19 is an artificial construct escaped from the laboratory, has no support in the analysis made from Andersen, and colleagues. Despite its affinity to the same ACE2 receptor, its affinity gained through a different binding domain, and the mutations it acquired are absolutely in line with the natural selection process.
Getting more samples of human CoV-19 and related viral samples from other animals would be the most clear-cut way to understand the mechanisms of its origin. Such knowledge will help us to prepare for future epidemics, which are probable to happen anyways, as we are an equal part of nature as viruses.
Original Article:
Andersen K, Rambaut A, Lipkin W, Holmes E, Garry R. The proximal origin of SARS-CoV-2. Nat Med. 2020.Edited by:
Massimo Caine , Founder and Director
We thought you might like
A new way to go gray
Nov 23, 2018 in Health & Physiology | 3.5 min read by Christopher R. Keys , Melissa HarrisVirus infection: may the (binding) force be with you?
Jan 11, 2019 in Microbiology | 4 min read by Martin Delguste , David AlsteensViruses are spilling over from managed honey bees to wild bumble bees
Mar 13, 2020 in Evolution & Behaviour | 4 min read by Samantha A. AlgerHow can a pathogen subvert honey bee social behaviors to increase its success?
May 21, 2021 in Evolution & Behaviour | 4 min read by Amy C. Geffre , Adam G. DolezalMore from Evolution & Behaviour
Cicada emergence alters forest food webs
Jan 31, 2025 in Evolution & Behaviour | 3.5 min read by Martha Weiss , John LillSize does not matter: direct estimations of mutation rates in baleen whales
Jan 29, 2025 in Evolution & Behaviour | 4 min read by Marcos Suárez-MenéndezThe Claws and the Spear: New Evidence of Neanderthal-Cave Lion Interactions
Jan 22, 2025 in Evolution & Behaviour | 3.5 min read by Gabriele RussoA deep-sea spa: the key to the pearl octopus’ success
Jan 20, 2025 in Evolution & Behaviour | 3.5 min read by Jim BarryFeisty fish and birds with attitude: Why does evolution not lead to identical individuals?
Aug 31, 2024 in Evolution & Behaviour | 3 min read by Lukas Eigentler , Klaus Reinhold , David KikuchiEditor's picks
Trending now
Popular topics


